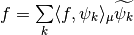Compare IW wavelets with classical wavelets: non linear approximation of signals¶
To play with the code you can run the Python File User_Example3.py
We now compare classical wavelets decomposition with intertwining wavelets studying non linear approximation of signals. The goal is to understand intertwining wavelets properties with respect to classical wavelets ones. In particular we want to check if intertwining wavelets have the classical properties we expect from a wavelet basis, especially the fact that if we threshold detail coefficients of a regular signal we can get a good approximation of our original signal.
To avoid confusion we have to say that there is no fast algorithm to compute intertwining coefficients (even if the toolbox iw was optimized as much as we could) and their computation is based on basic linear algebra.
We work with the file Torus_1024
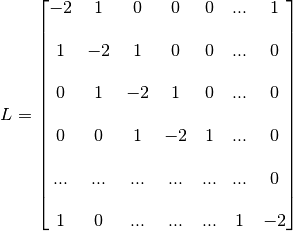
This graph connects each vertex with two neighbours. Its laplacian matrix is
We start with a quick description of the outputs of the main class IntertwiningWavelet and show how to process a signal.
Getting started¶
Load Python modules
The following Python modules should be useful.
scipy.sparse since we will use sparse matrices,
NumPy since we will process matrices and arrays,
matplotlib.pyplot for vizualisation
>>> import numpy as np
>>> import scipy.sparse as sp
>>> import matplotlib.pyplot as plt
Load the graph file
>>> from iw.data.get_dataset_path import get_dataset_path
>>> graph_file = get_dataset_path("tore_1024.g")
Start the instances of IntertwiningWavelet
>>> from iw.intertwining_wavelet import IntertwiningWavelet
>>> iw = IntertwiningWavelet(graph_file)
"The graph is reversible the pyramide algorithm....
can proceed"
>>> iw.pretreatment # To check if the graph has the required reversibility (symmetry)
True
Process the method
Here we choose to set the approximate cardinal of the set of approximation coefficients. Since we want to work with 5 levels of details in our classical wavelet basis, this amounts to have about 32 scaling coefficients.
>>> iw.process_analysis(mod='card', m=32)
# To have at most 32 approximation coefficients.
>>> print(iw.process_analysis_flag) # True if the decomposition process has been done.
True
>>> tab = iw.tab_Multires # Attribute with all the analysis and reconstruction structure
Process a signal¶
We will now compute the classical wavelet coefficients (CW coefficients) and intertwining ones (IW coefficients) of a signal.
Signal input: this is the classical model commonly used in the reference book by S. Mallat “A wavelet tour of signal processing”, Academic press.
>>> adr_signal = get_dataset_path("signal1D.mat")
>>> Sig = np.loadtxt(adr_signal) # download the signal
>>> n=np.size(Sig)
>>> Sig_iw = np.reshape(Sig,(1,n)) # reshape Sig in a 2d array to be able to run iw
Let us have a look on it
>>> plt.figure()
>>> plt.plot(Sig_iw[0,:]) # Watch out that Sig_iw is a 2d NumPy array
>>> plt.title('Original signal')
>>> plt.show()
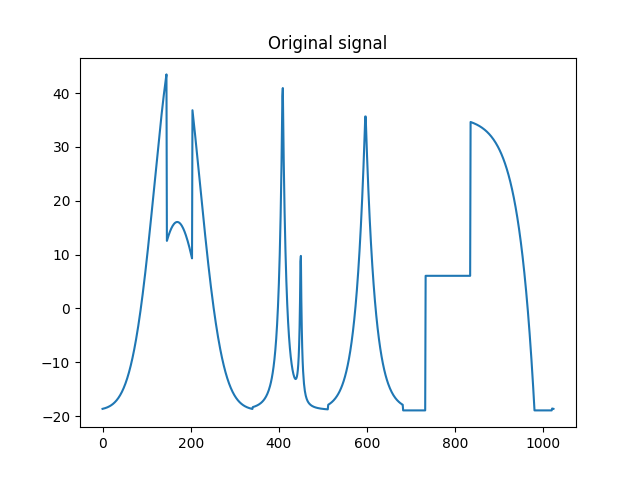
Original signal.¶
CW coefficients¶
Computation of classical wavelet coefficients
Use your favorite codes and your favorite wavelet basis to compute the classical wavelet coefficients (CW coefficients). Here we work with PyWavelets
>>> # import the PyWavelet toolbox
>>> import pywt
Choose your scaling function
>>> ond='db4'
Reshape the signal to have a simple array
>>> Sig_cw = Sig.copy()
>>> Sig_cw = np.reshape(Sig_cw,(n,))
Compute classical wavelet coefficients (CW coefficients) with 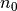 levels of detail coefficients.
levels of detail coefficients.
>>> n0=5
>>> coeffs_cw = pywt.wavedec(Sig_cw, ond, level=n0,mode = "periodization")
Let us visualize the CW coefficients.
>>> # the classical wavelet coefficients computed with Pywavelets are organized
# the other way around than IW wavelets.
>>> coeffs_vcw = np.concatenate(coeffs_cw[::-1])
>>> plt.figure()
>>> plt.plot(coeffs_vcw)
>>> plt.title('Classical wavelet coefficients with '+str(n0)+' levels' )
>>> plt.show()
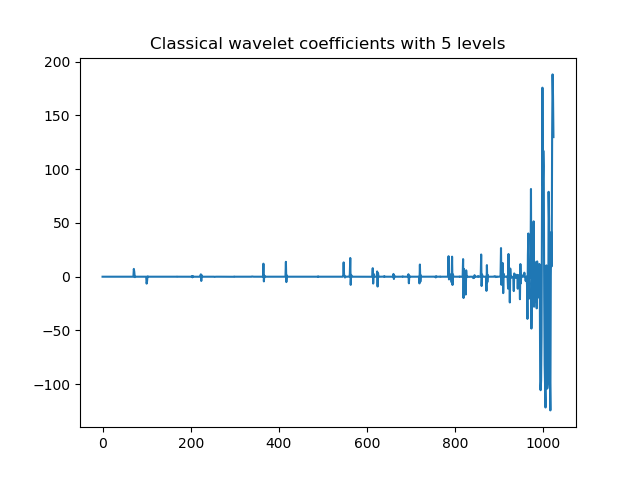
Classical wavelet coefficients with 5 levels.¶
IW coefficients¶
Computation of intertwining wavelet coefficients
This is done using the attribute of iw which is process_coefficients. The output is a 2d NumPy array, with possibly one line.
>>> coeffs_iw = iw.process_coefficients(Sig_iw)
>>> print(coeffs_iw.shape)
(1, 1024)
>>> plt.figure()
>>> plt.plot(coeffs_iw[0,:],'*') # Watch out that coeffs is a 2d NumPy array
>>> plt.title('Intertwining wavelet coefficients')
>>> plt.show()
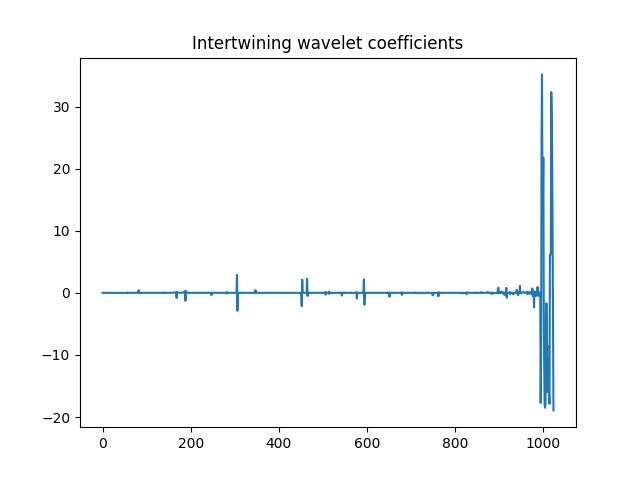
Intertwining wavelet coefficients.¶
Watch out that the Intertwining basis is not orthonormal, and especially the basis vectors are not normalized.
Organization of the coefficients:
The organization of the IW coefficients in the NumPy array coeffs_iw is as follows
coeffs_iw
with
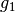 : the sequence of coefficients of the finest details level,
: the sequence of coefficients of the finest details level,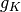 : the sequence of coefficients of the coarsest details level,
: the sequence of coefficients of the coarsest details level,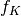 the sequence of scaling coefficients, or so called approximation coefficients.
the sequence of scaling coefficients, or so called approximation coefficients.
The attribute following_size of iw gives the number of coefficients in each layer
>>> levels_coeffs = np.asarray(iw.following_size)
>>> print(levels_coeffs)
[419 218 101 70 47 40 33 25 12 8 15 6 30]
In our example
the finest details level
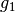 has 419 coefficients,
has 419 coefficients,the coarsest details level
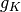 has 6 coefficients
has 6 coefficientswe have 30 approximation coefficients in
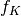 .
.
Remember our method is based on a random subsampling and thus the number of coefficients in each layer generally changes at each new run of iw. But we compute a basis and thus the total number of coefficients is always the total number of vertices in the graph.
Non linear approximation¶
We now compare the non linear approximations given by thresholding classical wavelets (CW wavelets) coefficients on one hand, and Intertwining wavelets (IW wavelets) coefficients on the other hand.
We first check the number of approximation CW coefficients. It should be 32 if we were not wrong !
>>> napprox=np.size(coeffs_cw[0])
>>> print(napprox)
32
With a classical wavelet basis we can also compute it using the following scheme
>>> nsig = np.floor(np.log2(n))
>>> na = np.floor(nsig-n0)
>>> napproxt = 2**(np.int(na)) # Compute the number of approximation coefficients
>>> print(napproxt)
32
Thresholding CW coefficients¶
We threshold the CW coefficients by keeping the 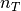 largest detail coefficients and reconstruct the thresholded signal. We will keep the approximation coefficients and will not threshold them.
largest detail coefficients and reconstruct the thresholded signal. We will keep the approximation coefficients and will not threshold them.
Let us write a function to compute the thresholded signal with the 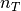 largest detail coefficients of the original signal
largest detail coefficients of the original signal
def non_linear_cw(sig,nT,coeffs_cw,ond):
# compute the number of the approximation coefficients
napprox = np.size(coeffs_cw[0])
# to get the numpy array which stores the coefficients
arr, sli = pywt.coeffs_to_array(coeffs_cw)
nl=np.size(arr) # total number of wavelet coefficients
# save the approximation which will not be thresholded.
arr_approx = arr[0:napprox].copy()
# we did not want to sort the approximation coefficients
arr[0:napprox] = np.zeros(napprox)
arg_T = np.argsort(np.abs(arr)) # we sort the detail coefficients
# we keep only the nT largest detail coefficients
arr[arg_T[0:nl-nT]] = np.zeros(nl-nT)
# to build the thresholded signal we restore the approximation
arr[0:napprox] = arr_approx
# we come back to the list of wavelet coefficients of the thresholded signal
coeffs_nT = pywt.array_to_coeffs(arr, sli)
# we reconstruct the signal with thresholded coefficients.
sig_nT = pywt.waverecn(coeffs_nT, ond, mode = "periodization")
return sig_nT
Compute the signal with 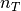 non vanishing detail coefficients and vizualize it.
non vanishing detail coefficients and vizualize it.
>>> nT = 100
>>> Sig_cw_nT = non_linear_cw(Sig_cw,nT,coeffs_cw,ond)
>>> plt.figure()
>>> plt.plot(Sig_cw_nT)
>>> plt.show()
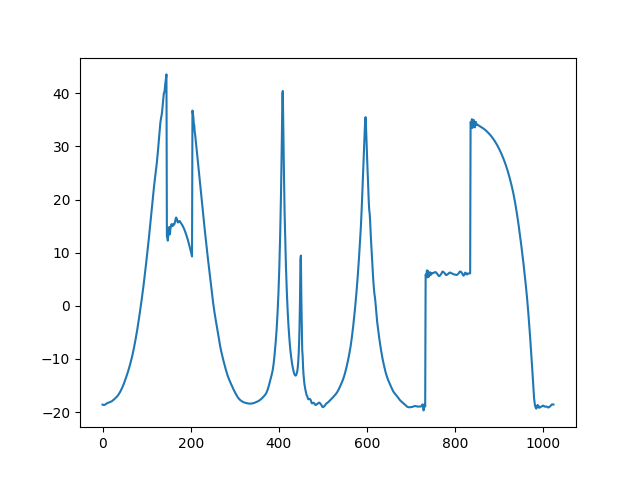
Thresholded signal: only 100 detail coefficients from the classical wavelet coefficients of the original signal are kept.¶
Thresholding IW coefficients¶
Normalization of intertwining wavelet coefficients
Our basis functions are not orthogonal and even not normalized. To threshold the coefficients an option is to normalize them in order to fix a way of comparing the size of the coefficients. There are several strategies one can choose.
We propose here the following scheme and we want to emphasize that one could choose another option.
Let us call 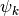 our basis functions. In other words the coefficients of a signal
our basis functions. In other words the coefficients of a signal 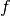 are computed through the formula
are computed through the formula 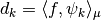 .
.
Recall that our setting allows to have a non uniform reversible measure 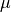 and we need to use the appropriate scalar product
and we need to use the appropriate scalar product
.
But if the Laplacian matrix 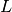 is as it is often the case symmetric then the scalar product
is as it is often the case symmetric then the scalar product 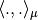 is the canonical one.
is the canonical one.
Since the functions of our basis ![\{\psi_k,k\in [0,n-1]\}](../_images/math/bcc6b8fa098ed4619711b1badaf0860f2d6e0c04.png) are non orthogonal but a linear independent system we can compute a family of functions
are non orthogonal but a linear independent system we can compute a family of functions 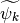 such that for each
such that for each 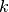 ,
, 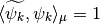 and for each
and for each 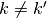 ,
, 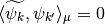 . This is called in general the dual system of the
. This is called in general the dual system of the ![\{\psi_k,k\in [0,n-1]\}](../_images/math/bcc6b8fa098ed4619711b1badaf0860f2d6e0c04.png) . The signal
. The signal 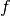 is given by
is given by
Our strategy is to compute 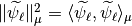 .
.
Then in a second step for each detail coefficient 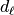 we will store
we will store 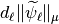 and sort it. Since we will not threshold the approximation coefficients we do not normalize the approximation coefficients.
and sort it. Since we will not threshold the approximation coefficients we do not normalize the approximation coefficients.
This is processed through the following function.
# to compute the mu-norm of the m first reconstruction functions
def norm_psi_tilde(m,mu):
# this matrix is the matrix of the iw-coefficients of the psi_tilde system
coeffs_dir = np.eye(m,n)
# (without the functions related to the approximation)
# compute the psi_tilde family
#(without the approximation reconstruction functions)
psi_tilde = iw.process_signal(coeffs_dir)
# compute the collection of norms of the psi_tilde vectors
norm_psi_tilde = np.linalg.norm(psi_tilde*np.sqrt(mu),axis=1)
return norm_psi_tilde
We apply this function and compute 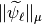 for all detail functions (
for all detail functions (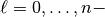
napprox).
>>> n = np.size(coeffs_iw) # This yields the total number of coefficients
>>> # to get the sequence of coefficient's number by level
>>> levels_coeffs = np.asarray(iw.following_size)
>>> # to get the number of approximation coefficients
>>> napprox = levels_coeffs[tab.steps]
>>> # We want to compute all the norms of the detail functions
>>> m = n-napprox
>>> # iw gives the reversible measure which is the uniform measure if L is symmetric
>>> mu = np.asarray(iw.mu_initial)
>>> mu_r = np.reshape(mu,(1,n))/np.sum(mu) # we work with a row vector.
>>> n_psi_tilde = norm_psi_tilde(m,mu_r)
Let us visualize it. We can see clearly that our functions are not normalized.
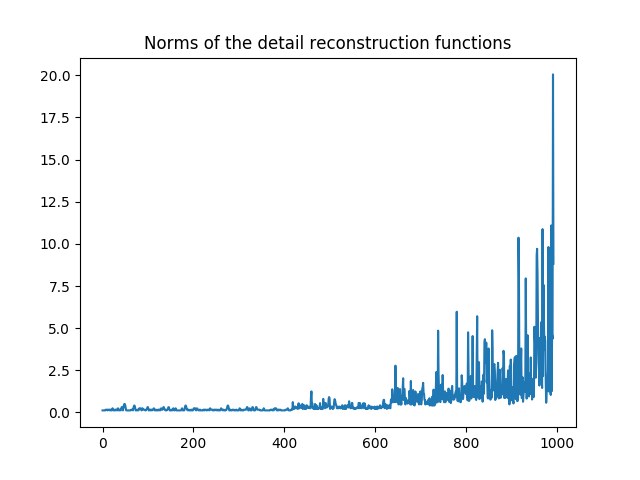
Norms 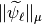 for all detail reconstruction functions.¶
for all detail reconstruction functions.¶
Threshold the IW coefficients and compute the thresholded signal
We have now to compute 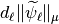 and sort them before computing the thresholded signal. All of this is done using the following function.
and sort them before computing the thresholded signal. All of this is done using the following function.
def non_linear_iw(sig,nT,coeffs_iw,n_psi_tilde):
n = np.size(coeffs_iw) # This yields the total number of coefficients
# to get the sequence of coefficient's number
levels_coeffs = np.asarray(iw.following_size)
# to get the number of approximation coefficients
napprox = levels_coeffs[tab.steps]
# compute the number of the approximation coefficients
m=n-napprox
coeffs_iwT = coeffs_iw.copy()
coeffs_iwn = coeffs_iwT[0,0:m].copy()*n_psi_tilde
# we sort the detail coefficients
arg_iwT = np.argsort(np.abs(coeffs_iwn))
# we keep only the nT largest detail coefficients
coeffs_iwT[0,arg_iwT[0:m-nT]] = np.zeros((1,m-nT))
# Reconstruct the signal
sig_nT=iw.process_signal(coeffs_iwT)
return sig_nT
Compute the signal with 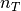 non vanishing detail coefficients and vizualize it.
non vanishing detail coefficients and vizualize it.
>>> nT = 100
>>> Sig_iw_nT = non_linear_iw(Sig_iw,nT,coeffs_iw,n_psi_tilde)
>>> plt.figure()
>>> plt.plot(Sig_iw_nT)
>>> plt.show()
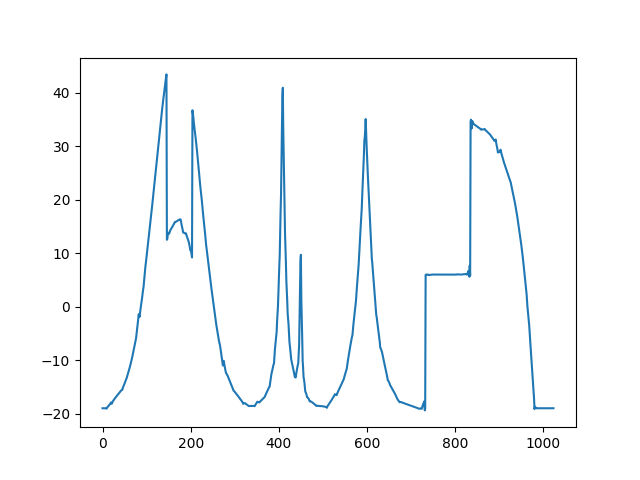
Thresholded signal: only 100 detail coefficients from the IW coefficients of the original signal are kept.¶
We have another way to compute 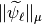 . This is done using the reconstruction operators as described in Reconstruction operators. We take now the opportunity to explore more in details the analysis and reconstruction operators computed by
. This is done using the reconstruction operators as described in Reconstruction operators. We take now the opportunity to explore more in details the analysis and reconstruction operators computed by iw.
Analysis and reconstruction¶
The main attribute tab_Multires of iw contains the sequence of subgraphs and contains also the basis. Recall that we have
>>> tab = iw.tab_Multires # Attribute with all the analysis structure
The variable tab is a MemoryView which has three attributes
>>> print(tab)
<iw.multiresolution.struct_multires_Lbarre.Tab_Struct_multires_Lbarre object at 0x7f3186287e30>
The attribute steps: it is the number of decomposition levels
>>> tab.steps # To get the number of decomposition levels
The attribute Struct_Mres_gr: it is the sequence of subgraphs which is as well a MemoryView.
To know more on the structure of subgraphs and how to get access to the information go back to the tutorial Graphs and subgraphs.
The third attribute of tab is Struct_Mana_re. It stores the analysis operator to compute the wavelet coefficients and the reconstruction operators to compute a signal given its coefficients. It is again a MemoryView object
>>> basis = tab.Struct_Mana_re
>>> print(basis)
<MemoryView of 'ndarray' object>
>>> k = 0 # To access to the operators at the finest level (finest scale)
>>> a0 = basis[k] # To access to the operators at level k
The attributes of basis store all the operators needed to analyse signals, ie. to compute wavelets coefficients, and the operators to reconstruct the signals given coefficients. To fix the notations recall the multiresolution scheme to analyse a signal 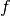
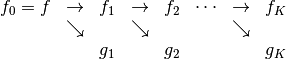
Thus at each level 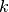 we have access to the following matrices
we have access to the following matrices
the analysis matrix
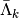 computes the approximation coefficients
computes the approximation coefficients 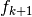 from
from 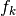 , i.e
, i.e
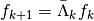
the matrix
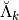 computes the detail coefficients
computes the detail coefficients 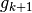 from
from 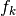 , i.e
, i.e
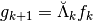
the reconstruction operators
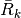 and
and 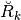 are such that
are such that
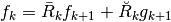
Recall that the diacritical accents in ancient greek  and
and  are marked with respectively a bar and a breve.
are marked with respectively a bar and a breve.
Analysis operators¶
Let us have a closer look at the first level. We expect if we compute 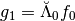 to recover the finest detail coefficients of our signal.
to recover the finest detail coefficients of our signal.
The analysis detail operator 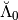 is sorted as a MemoryView in the attribute
is sorted as a MemoryView in the attribute Lambdabreve of basis at level 0.
>>> print(a0.Lambdabreve)
<MemoryView of 'ndarray' object>
>>> Lambdabreve_0 = sp.coo_matrix((a0.Lambdabreve,(a0.rowLambdabreve,a0.colLambdabreve)),
shape=(a0.shape0Lambdabreve, a0.shape1Lambdabreve))
>>> Lambdabreve0 = Lambdabreve_0.toarray()
Let us check its size. It has the same number of columns as the graph and the same number of rows as the finest detail part
>>> print(Lambdabreve0.shape) # Shape of the matrix Lambdabreve0
(419, 1024)
>>> print(levels_coeffs[0]) # Number of finest detail coefficients
419
We should recover the finest detail coefficients through the product 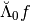 .
.
>>> # Remember our signal is f=Sig and we take the numpy vector version.
>>> g1=Lambdabreve0@Sig_cw
>>> # Extract the finest detail part from the IW coefficients
>>> coeffs_g1=coeffs_iw[0,0:levels_coeffs[0]]
>>> # Check that there are the same up to very small computation errors
>>> print(np.linalg.norm(coeffs_g1-g1))
1.2850453296247093e-14
If we want to recover 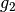 we need to compute
we need to compute 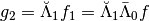 and we can do the following computation.
and we can do the following computation.
The matrix 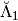 is sorted as a MemoryView in the attribute
is sorted as a MemoryView in the attribute Lambdabreve of basis at level 1. The matrix 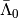 is sorted as a MemoryView in the attribute
is sorted as a MemoryView in the attribute Lambdabarre of basis at level 0.
>>> a1 = basis[1] # Compute the operators at the further step
>>> # Compute the matrix Lambdabreve1
>>> Lambdabreve_1 = sp.coo_matrix((a1.Lambdabreve,(a1.rowLambdabreve,a1.colLambdabreve)),
shape=(a1.shape0Lambdabreve, a1.shape1Lambdabreve))
>>> Lambdabreve1 = Lambdabreve_1.toarray()
>>> # Compute the matrix Lambdabarre0
>>> Lambdabarre_0 = sp.coo_matrix((a0.Lambdabarre,(a0.rowLambdabarre,a0.colLambdabarre)),
shape=(a0.shape0Lambdabarre, a0.shape1Lambdabarre))
>>> Lambdabarre0 = Lambdabarre_0.toarray()
>>> # Remember our signal is f=Sig and we take the numpy vector version.
>>> g2=Lambdabreve1@(Lambdabarre0@Sig_cw)
>>> # Extract the finest detail part from the IW coefficients computed with IW
>>> coeffs_g2=coeffs_iw[0,levels_coeffs[0]:levels_coeffs[0]+levels_coeffs[1]]
>>> # Check that there are the same up to computer precision
>>> print(np.linalg.norm(coeffs_g2-g2))
1.3736784588236522e-14
Reconstruction operators¶
At the first level we can have a look at 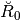 . It is is stored in the attribute
. It is is stored in the attribute Reconsbreve of basis at level 0.
>>> Rbreve_0 = sp.coo_matrix((a0.Reconsbreve,(a0.Recons_row_breve,a0.Recons_col_breve)),
shape=(a0.Recons_shape0_breve, a0.Recons_shape1_breve))
>>> Rbreve0 = Rbreve_0.toarray()
Let us check its size. It has the same number of rows as the graph and the same number of columns as the finest detail part
>>> print(Rbreve0.shape) # Shape of the matrix Rbreve0
(1024, 419)
>>> print(levels_coeffs[0]) # Number of finest detail coefficients
419
Remark that since 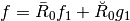 the columns vectors of
the columns vectors of 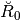 are exactly the functions
are exactly the functions 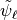 corresponding to the finest detail reconstruction functions. This can be checked by an easy computation either numerically and/or theoritically and is left as an exercize to the reader.
corresponding to the finest detail reconstruction functions. This can be checked by an easy computation either numerically and/or theoritically and is left as an exercize to the reader.
We already computed 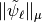 in the previous section, and extract the contribution of the finest detail level. We check now that if we compute the
in the previous section, and extract the contribution of the finest detail level. We check now that if we compute the 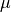 -norms of the columns of the reconstruction operator of
the finest detail level we obtain the same result.
-norms of the columns of the reconstruction operator of
the finest detail level we obtain the same result.
>>> mu_r=np.reshape(mu_r,(n,1)) # We need a column vector since we compute norms of columns vectors
>>> # Compute the collection of mu-norms of the column vectors of Rbreve0
>>> norm_Rbreve0 = np.linalg.norm(Rbreve0*np.sqrt(mu_r),axis=0)
>>> nd1=norm_Rbreve0.size
>>> # difference between the results for the finest detail reconstruction functions
>>> # given by the two methods
>>> plt.figure()
>>> plt.plot(n_psi_tilde[0:nd1]-norm_Rbreve0)
>>> plt.show()
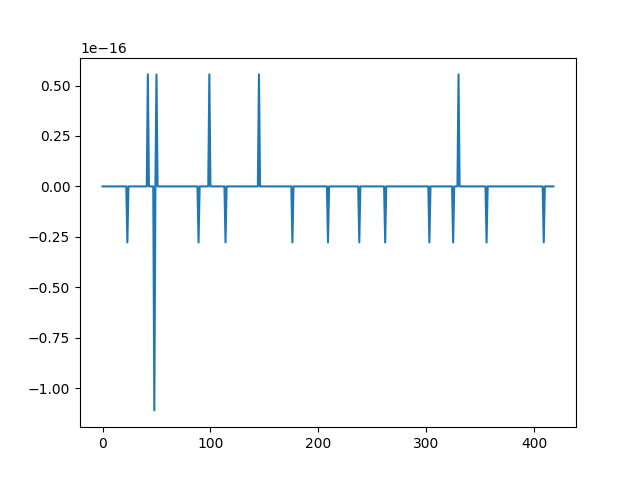
The two methods for computing the norms of the 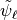 at the finest detail level yield the same result up to computer precision.¶
at the finest detail level yield the same result up to computer precision.¶
We can go on if we want and compute the norms of the columns vectors of  to compute all the
to compute all the 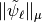 . We leave it to the reader !
. We leave it to the reader !
![=[[g_1,g_2,\dots,g_K,f_K]]](../_images/math/be807460c73844e7df6dafabd2743e53207252ba.png)